Metadata Configurator#
A free to use FAIR Data Station is available at https://fairds.fairbydesign.nl or setup your own.
The Metadata Configurator allows you to configure your own metadata templates by selecting metadata fields and sheets that are of relevance for your research. The information can be ultimately transformed into a Linked Data file with the Metadata Validator which can be used for automatic processing of data sets, publishing and exploratory queries over a multitude of studies.
This will give an interface that looks like this:
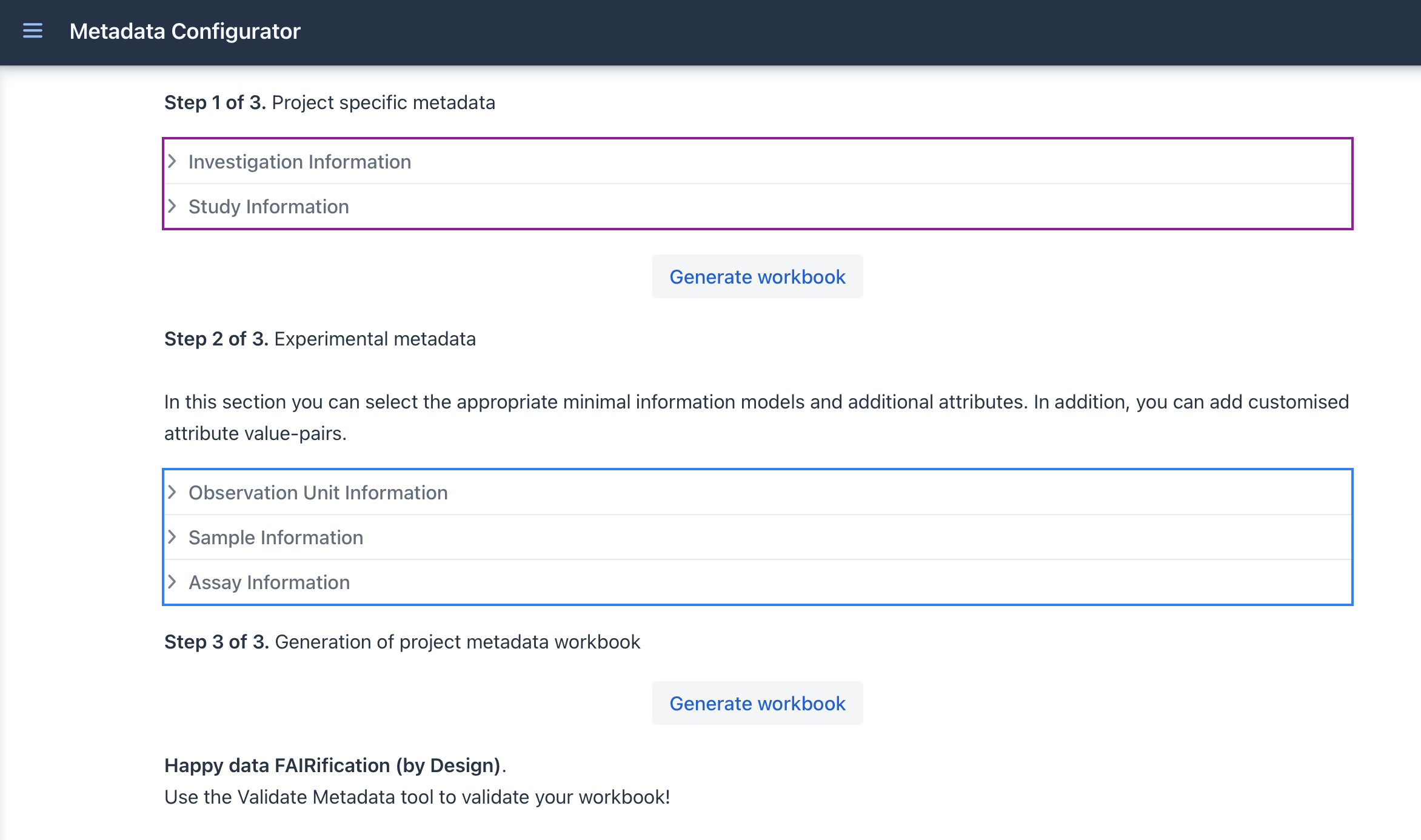
The Metadata Configuration Web Form is divided in to different segments; Investigation,Study, etc.. You can see the relations between these segments in Figure below.
An explanation of each segment can be found on this page. And also what we expect it to contain.
After the creation of the template you can click on the GENERATE EXCEL button to create your project metadata file.
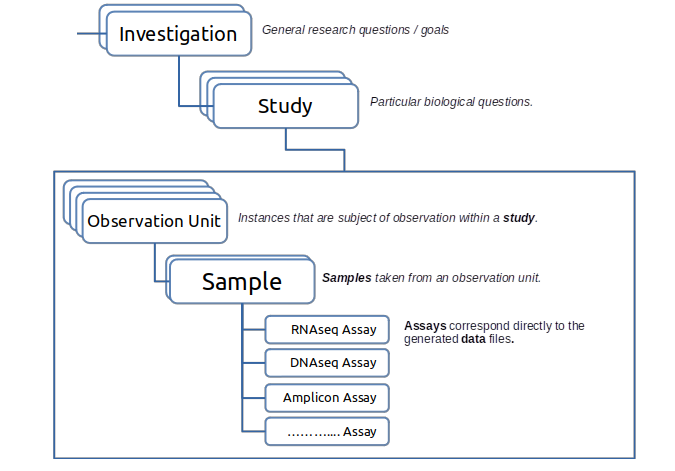
THE FAIRDS Metadata Structure
Investigation#
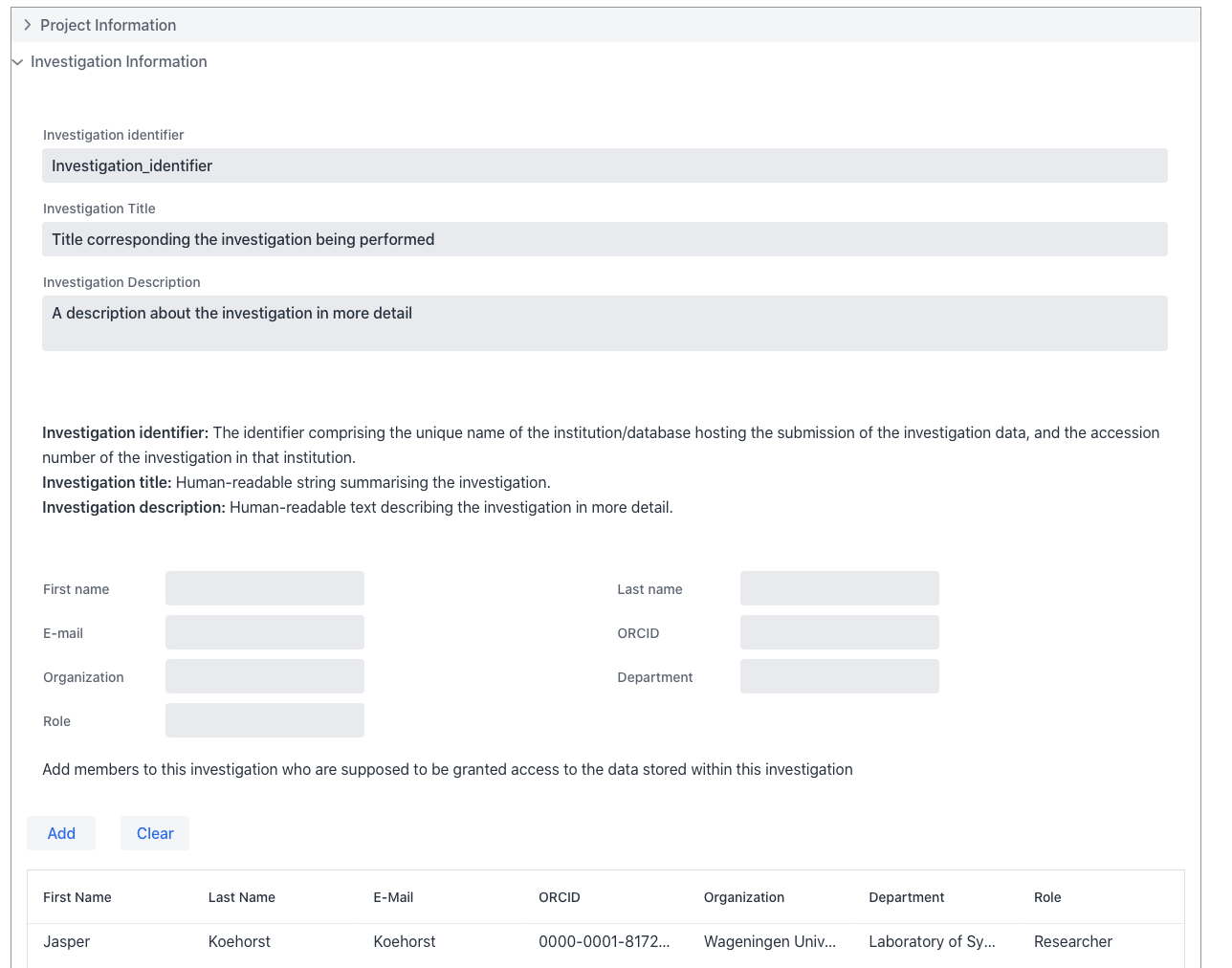
Investigations are defined here as research questions within the the specified project. The Web form is designed to enter a single investigation.
Investigations is the root of the metadata structure. It is the highest level of the metadata structure.
Example of the Investigation section
Study#
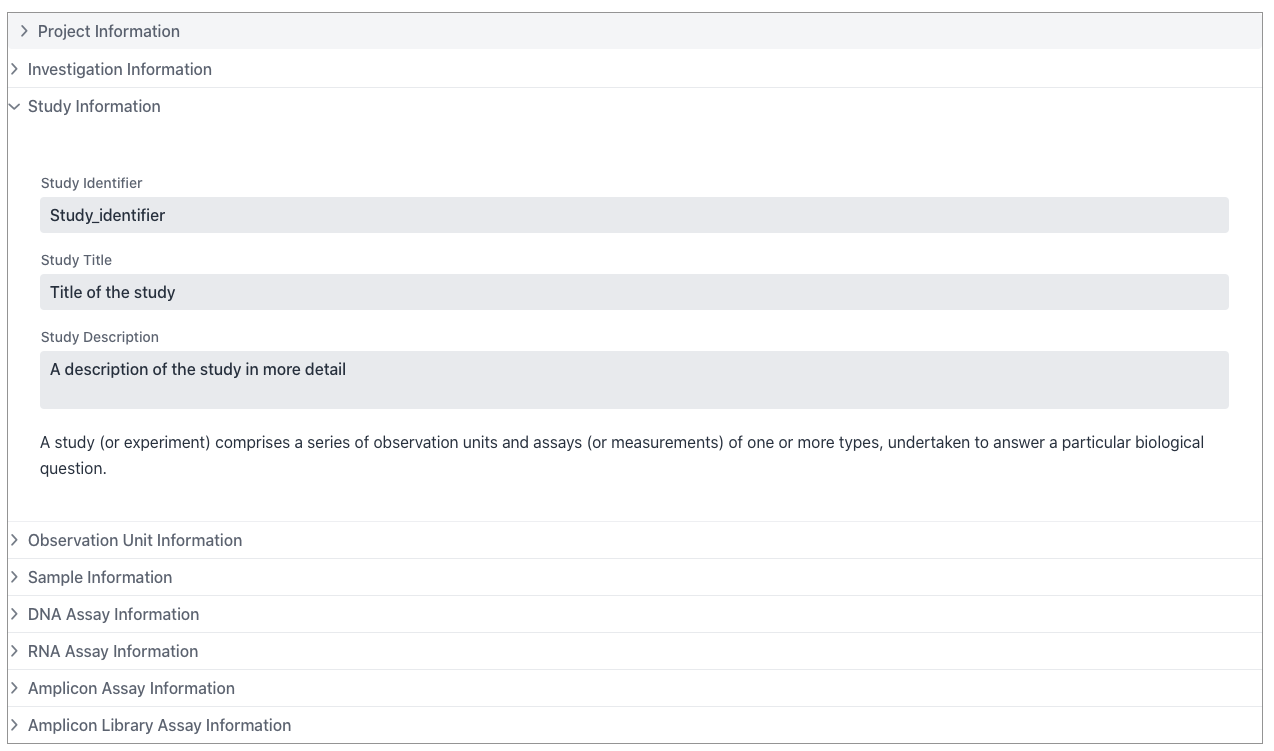
A study comprises a series of observation units (see below) and assays or measurements of one or more types, undertaken to answer a particular biological question within it’s investigation.
Every Study has an associated Investigation
Example of the Study section
Observation Unit#
Observation units are objects that are subject to instances of observation and measurement.
Every Observation Unit is associated with a Study
For example in a study each entity patient, plant, animal, bioreactor or area studied is one observation unit. When studying 50 different animals, each animal should become one observation unit. Additional packages will be released that support different observation unit types such as bioreactors, patients, animals, fields or oceans.
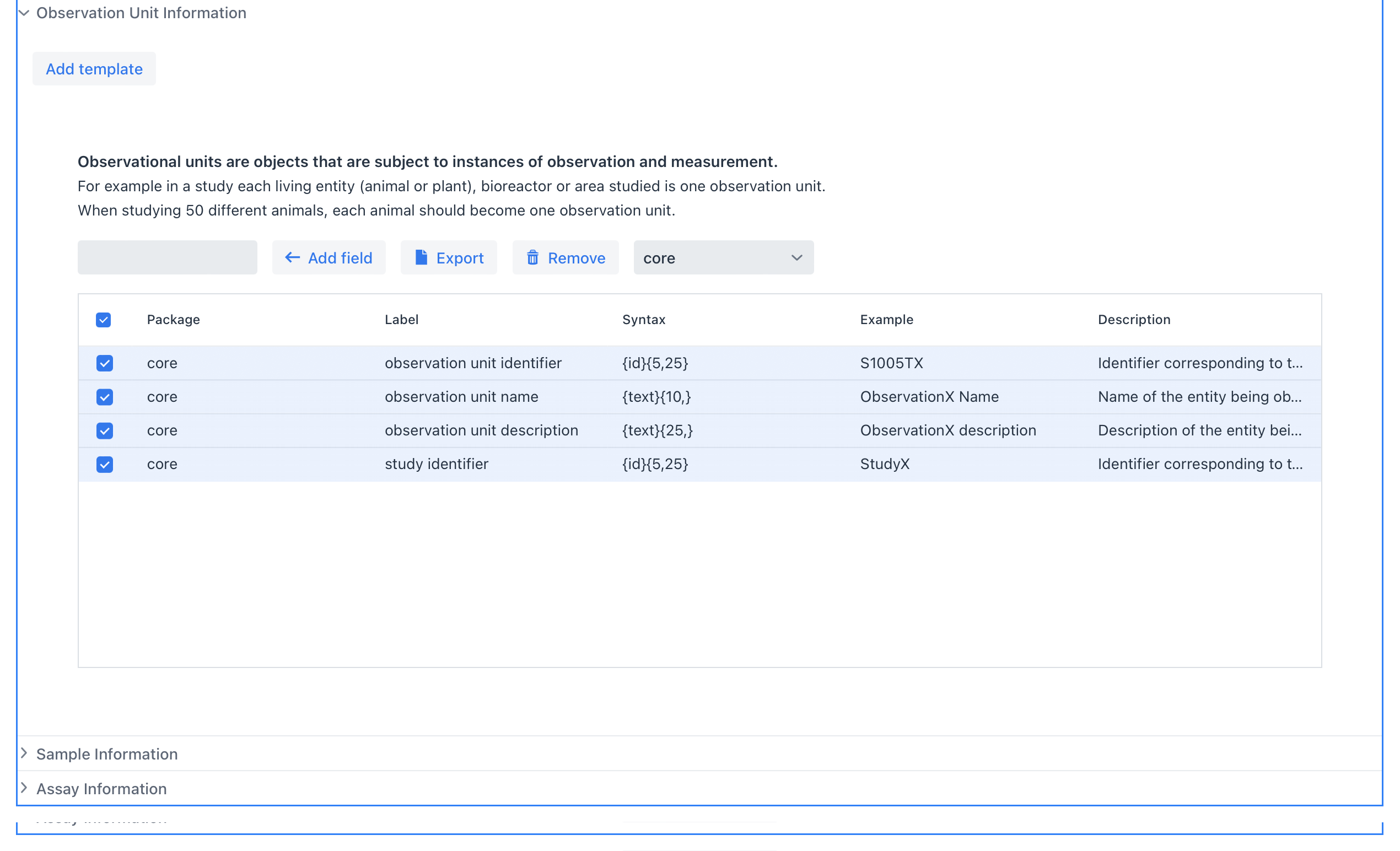 Example of the Observation Unit section
Example of the Observation Unit section
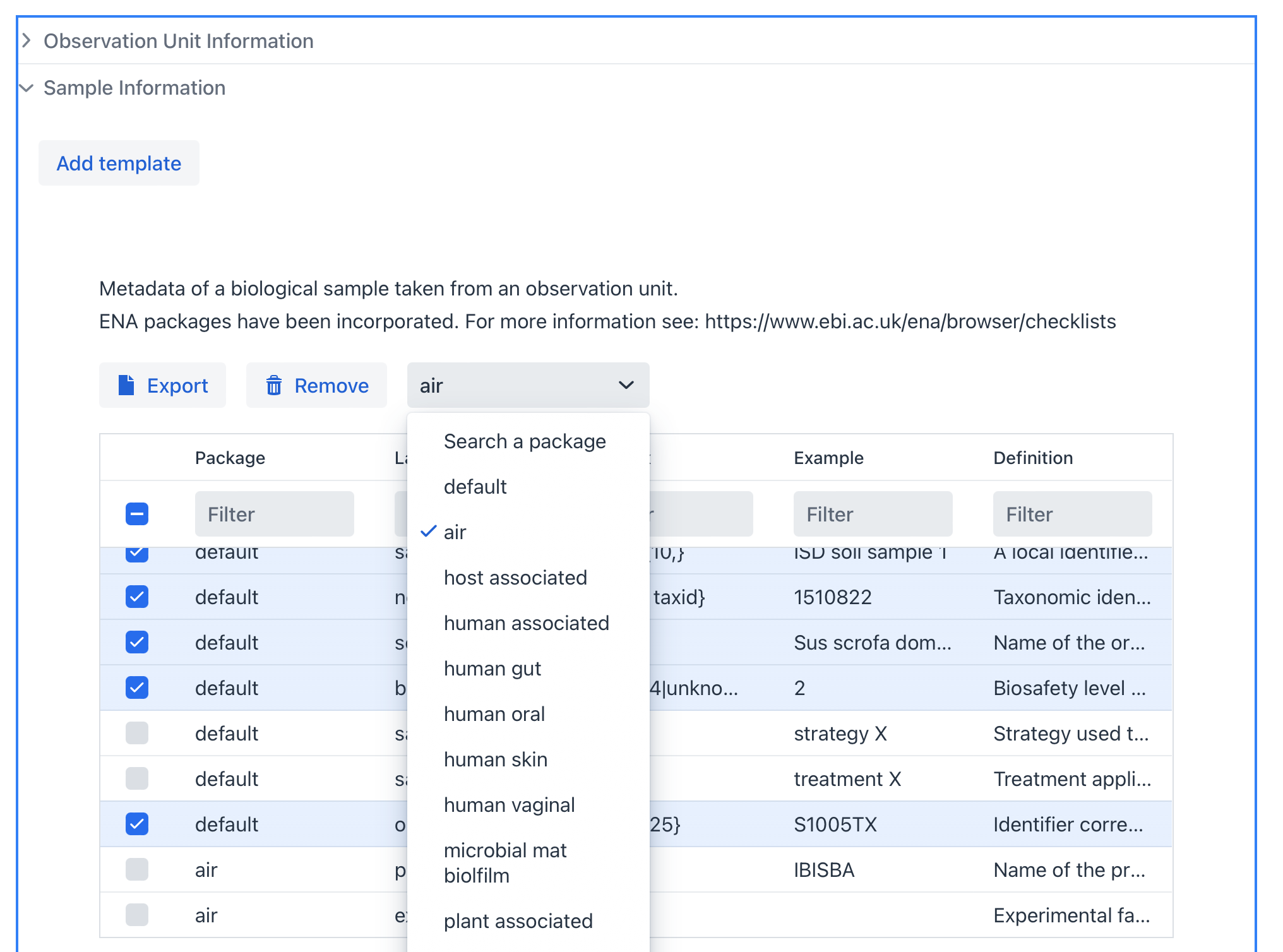
Sample#
A Sample is taken from an Observation Unit that can potentially be processed further to acquire data from. A multitude of samples can be taken from one or more observation units when for example performing time-scale experiments or sampling from different regions of the observation unit.
A varity of sample packages are available and are based on the MIxS standard (https://w3id.org/mixs). Each package has its own obligatory fields and varying additional fields depending on the sample type.
It also possible to have different sample types belonging to the same Observation Unit. You can add multiple sample types by clicking on the Add template button.
Example of the Sample section
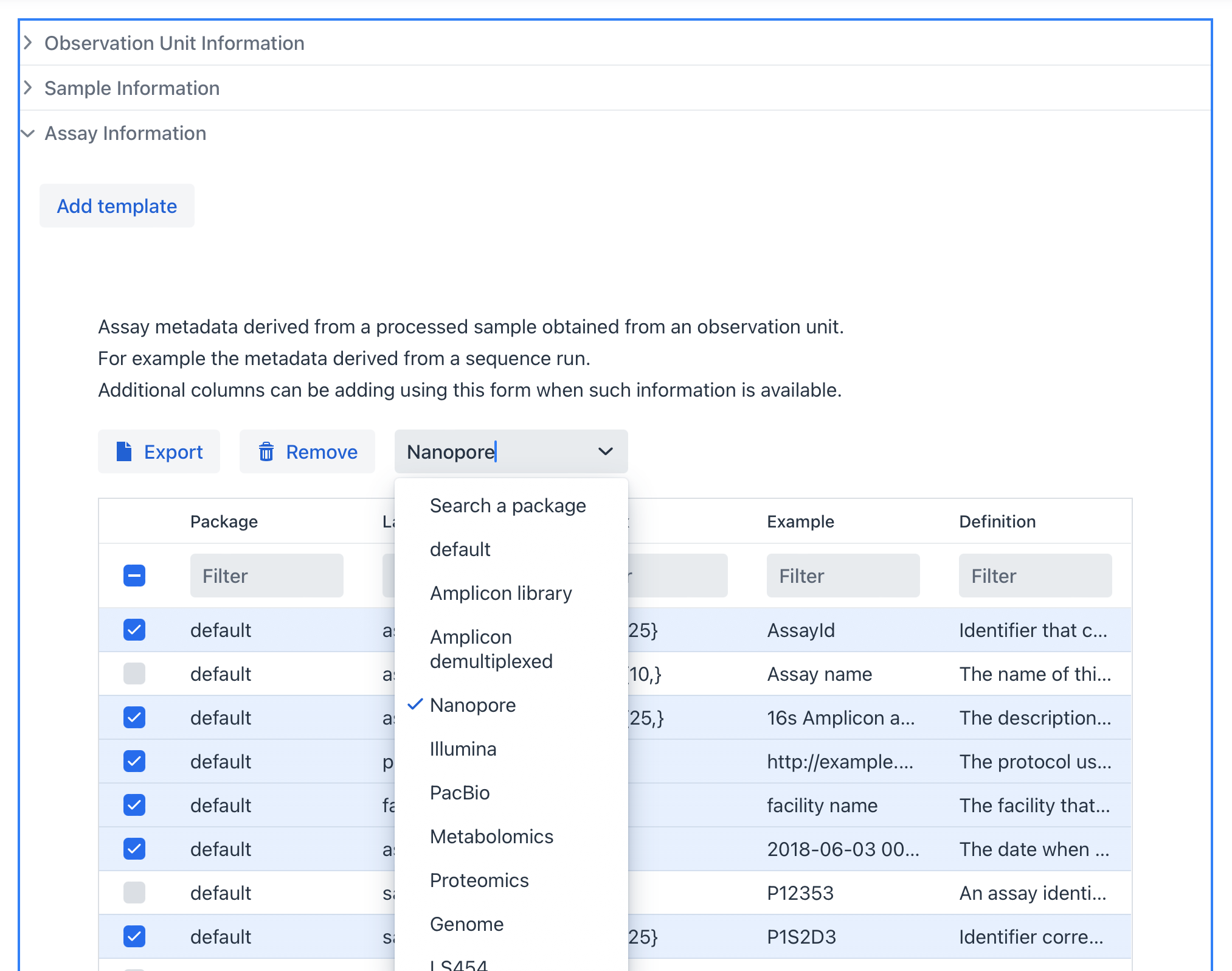
Assays#
An Assay corresponds to the data (for example a sequencing run) that was performed on a sample obtained from its observation unit.
When you have multiple types of assays, for example DNA and RNA, you can enable both sheets and appropriate rows. In addition, columns can be adding using this form when such information is available.
Example of the DNA assay section:
Generate#
When you have finished filling in the investigation and study information, selected the right observation units, samples and assays you can hit the generate button to create the metadata file.
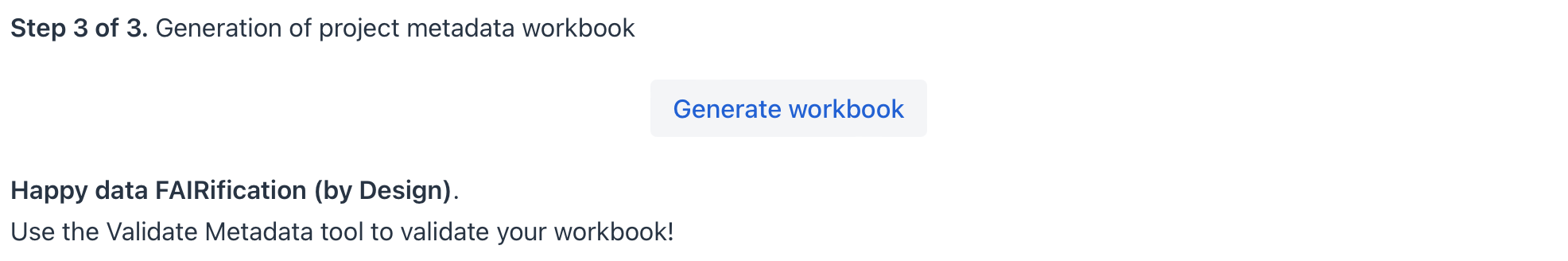
If you realise later on you missed a package for one of the different experimental metadatasets you can always download them individually using the Export button associated with each metadata set.
