FAIR Data Management#
The ‘FAIR Guiding Principles for scientific data management and stewardship’ are built upon the use of machine-actionable metadata to find, access, interoperate, combine, and directly reuse data with minimal human intervention.
To improve the quality of the reported data and to maximise the potential for reuse, the set of metadata must be sufficient to allow for unambiguous interpretation of the associated data. In the Life Sciences, Minimum Information standards are used for metadata management, which consist of two parts. Firstly, for each assay and associated data type there is a community accepted checklist of reporting requirements. Secondly, an obligatory data format is used for reporting essential metadata to ensure machine-actionability.
FAIR BY DESIGN#
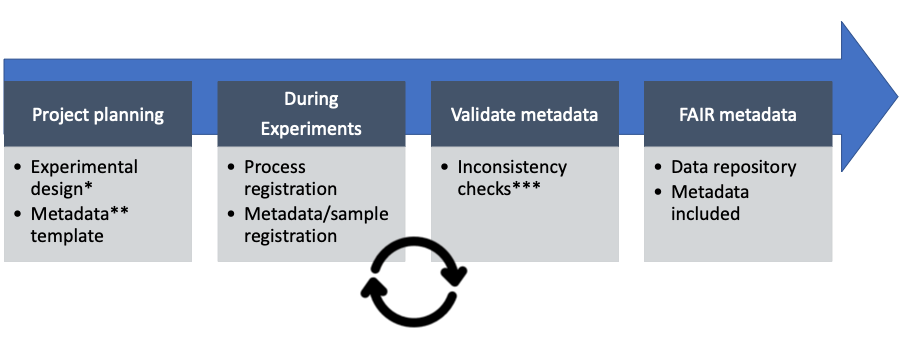
Working in a FAIR by Design manner ensures projects are Findable, Accessible, Interoperable and Reusable from the start. Throughout all the stages of the project FAIR principles are applied to improve the quality of your research.
This process is initiated by project planning and experimental design using the FAIR Data Station https://fairds.fairbydesign.nl. In which you can record all metadata important for your research. The information is then transformed into a Linked Data file which can be used for automatic processing of datasets, publishing and exploratory queries over a multitude of studies.
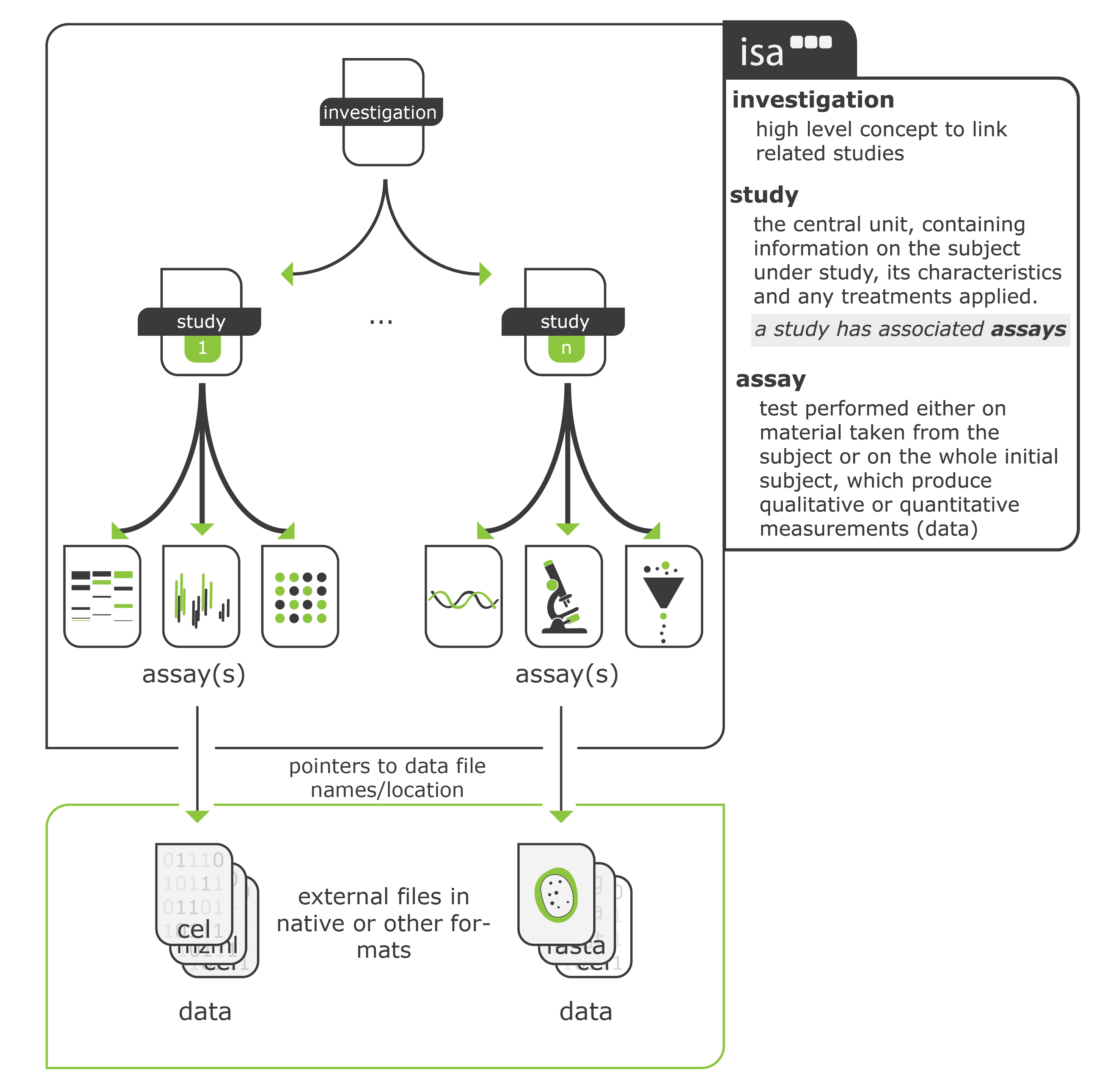
Role of ISA#
The core of our Linked metadata system is based on the ISA format (http://isa-tools.org). ISA (Investigation, Study and Assay) is a metadata framework used to manage a diverse set of experiments from the life, environmental and biomedical sciences. The Investigation (the project context), Study (a unit of research) and Assay (analytical measurements) concepts are incorporated and expanded with other standards such as JERM, MIAPPE and MIxS.
Ontologies#
A set of ontologies are combined and used to support the metadata model (http://git.wur.nl/unlock/ontology). This model is based on the ISA structrure in which the URI’s from ISA have been mapped to the Just Enough Results Model. Through JERM the Project class was added to improve the alignment with the FAIRDOM Hub and to improve the categorisation of multiple investigations under one funding agency (the project).
In biological studies, samples play a key role and in almost all research projects more then one subject, entity or environment is being analysed. The addition of Observation Unit from the MIAPPE ontology solves the problem of the missing association between sample and study.

API#
The ontology is written in a combination of OWL and ShEx enabling Empusa to generate a JAVA API for integration purposes. The API available at http://download.systemsbiology.nl/#/unlock/ is used to create, manage and query the RDF datasets generated throughout various applications. First and foremost the ontology is used by default in the FAIR Data Station, accessible at https://fairds.fairbydesign.nl to validate and generate structured RDF data from metadata excel sheets.
The generated RDF datasets are then used to drive the computational work using the metadata information available in the RDF datasets and the Common Workflow Language for processing raw data to information.
